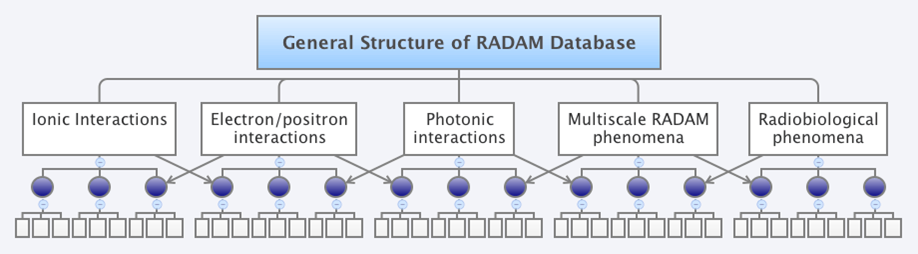
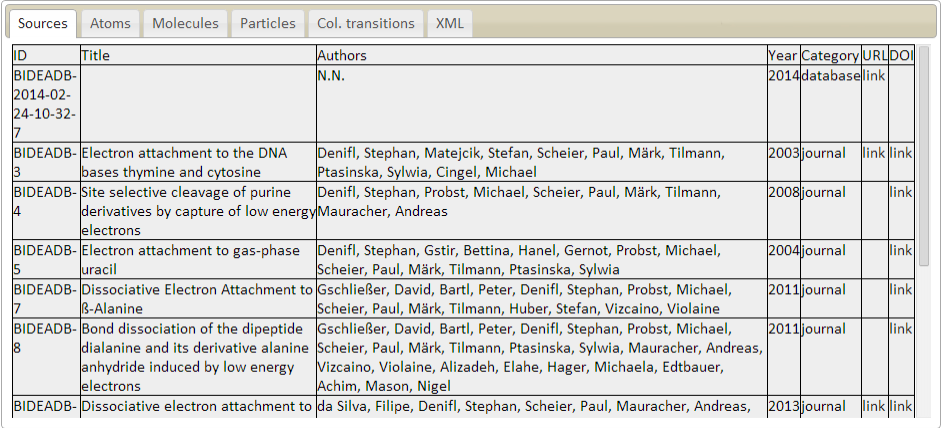
Sources/bibliographic references
ID - source identifier
Name - name of the journal, database or other source
Title - title of the paper
Authors - list of authors of the paper
Year - publication year
Category - type of the source (journal/database)
URL - link to the source
DOI - link to the source with specified DOI
Description: Sources table describes the list of external sources of the information in the database. Each
entry in the database must reference published paper or external database as a source of information.
EnergyLoss: (LETCurves)
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
SpeciesRef - reference to specie description
InitialEnergy - initial energy of the particles
BraggPeakPosition - position of the Bragg peak
BraggPeakWidth - width of the Bragg peak in x
BraggPeakEnergyWidth - width of the Bragg peak in T
DataSets - arbitrary set of tables and plots
envRef - reference to environment description
Description: Linear Energy Transfer (LET) [eV/nm] :: energy absorbed by the medium per unit length
of ion's path. LET depend on the medium and on the projectile. The main defining factors are the
charge of the projectile, its velocity, and electron density of the medium. More detailed information
includes specific information on states of the target. LET dependence is characterized by the Bragg peak,
proximal plateau, and distal tail. For a given ion and medium, the position of the Bragg peak is largely
defined by the initial energy of the ion; its height by the ion's charge. Since the charge changes as ions
slow down (charge transfer effect) the height is proportional to the square of the effective charge that
depends on velocity. For a single ion the Bragg peak is only defined by the above data. For the whole ion
beam, the position of the Bragg peak is averaged over all ions, which have different scattering histories.
Therefore the Bragg peak is significantly lower and wider (energy straggling effect).
StragglingAndBeams
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
SpeciesRef - reference to specie description
StragglingSigma - value of longitudinal straggling deviation
AngularSigma - value of lateral straggling deviation
DataSets - arbitrary set of tables and plots
envRef - reference to environment description
For the whole ion beam, the position of the Bragg peak is averaged over all ions, which have different
scattering histories. Therefore the Bragg peak is significantly lower and wider (energy straggling effect).
This effect is taken into account by the assumption that the widening of the Bragg peak is Gaussian.
The width of this Gaussian is stored below as StragglingSigma for different media and projectiles.
AngularSigma is a similar parameter for the lateral widening of the beam.
ThermomechanicalDamage
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
SpeciesRef - reference to specie description
InitialEnergy - initial energy of the particles
DataSets - arbitrary set of tables and plots, e.g. Dependence of temperature in the thermal spike as a function of radius and time.
envRef - reference to environment description
Secondary electrons deposit their energy in a very small region (radius of 1-1.5 nm) around the ion's
path. As a result the temperature rapidly increases in this region. This temperature dependence on
the distance from ions path and time after traverse has been predicted for a number of scenarios in
[ThermalSpikes]. As a result of thermal spikes, the pressure in the small region near the pass rapidly
increases as well and this becomes the source of cylindrical shock waves predicted in [ShockWaves]. The
direct effect of shock waves on biomolecules have been studied; and the probability of strand breaks
due to rupture of covalent bonds by stresses has been evaluated using molecular dynamics simulations.
This probability depends on LET and the proximity of biomolecules to the ion's path. Another effect of
shock waves relevant to biodamage is the radial transport of reactive species formed in the vicinity to
the path. This transport is more effective than diffusion.
ComplexDielectricFunction
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
SpeciesRef - reference to specie description
DataSets - arbitrary set of tables and plots
envRef - reference to environment description
Complex dielectric function is used to calculate cross sections of ionization for a variety of organic
molecules and biomolecules. The energy loss-function (ELF) is defined as the negative imaginary part of
the inverse dielectric function (they can be stored in the database for a number of molecules as figures
or parameterizations). Singly differential cross section obtained using the ELF for different species can
be stored as well as the total cross sections depending on the energy of projectiles (protons).
TransportOfSecondaryParticles
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
SpeciesRef - reference to specie
DiffusionCoefficient - value of the diffusion coefficient in a given medium
MeanFreePath - value of mean free path in a given medium (only for radicals and solvated electrons
DataSets - arbitrary set of tables and plots
envRef - reference to environment description
More than 80% of secondary electrons ejected by ions have energies less than 50 eV. At these
energies, the cross sections are mostly isotropic. Therefore the random walk method can be used
for the description of their transport. The random walk method is similar to the diffusion. Therefore,
the diffusion coefficients for electrons are main parameters describing this transport. The diffusion
coefficients are energy-dependent and given by D=lv/6, where l is the mean free path and v is the
electron's velocity. Therefore, the dependence of the mean free paths (both elastic and inelastic) are
stored here. For other species, such as free radicals, solvated electrons, and other secondary particles,
the transport is also explained by diffusion and the diffusion coefficients are stored here.
In addition the mean penetration lengths for all species including electrons (depending on their initial
energies) can be stored here.
ChemReactionsSecondaries
MethodRef - reference to a method description
Comments - arbitrary comments
SourceRef - bibliographic reference to a source of the data
MaximalReactionDistance - maximal reaction distance value
ReactionRateConstant - value of the reaction rate constant
Reactant - references to species description
IntermediateState - list of intermediate states
Product - references to species description
DataSets - arbitrary set of tables and plots
envRef - reference to environment description
These are the main parameters of chemical reactions relevant for the secondary particle propagation.
Molecular state
Molecule Ref. - Reference to a molecule
State energy - Energy of the state
Energy origin - Reference to a ground state
ElecStateLabel - The label of the electronic state: X, a, A, b, etc...
V1 - The vibrational quantum number, v1
V2 - The vibrational quantum number, v2
L2 - The quantum number, l2, associated with the vibrational angular momentum of the nu2 bending mode
V3 - The vibrational quantum number, v3
J - The rotational quantum number, J, associated with the total angular momentum excluding nuclear spin
I - The quantum number associated with the total nuclear spin angular momentum resulting from the coupling of two individual nuclear spin angular momenta: I = I1 + I2
F1 - The quantum number, F1, associated with the intermediate angular momentum due to coupling the rotational angular momentum with one nuclear spin. F1 may not be a good quantum number.
F2 - The quantum number, F2, associated with the intermediate angular momentum due to coupling the rotational angular momentum with one nuclear spin. F2 may not be a good quantum number.
F - The quantum number, F, associated with the total angular momentum including nuclear spin
r - A ranking index for states of the same symmetry that can't be or haven't been differentiated any other way: r=1,2,...
Parity - Total parity with respect to inversion through the molecular centre of mass in the laboratory coordinate system
KronigParity - Kronig parity ('e' or 'f')
asSym - Symmetry of the rovibronic wavefunction for diatomic molecules with a centre of inversion: a or s such that the total wavefunction including nuclear spin is symmetric or antisymmetric under permutation of the identical nuclei, according to whether they are bosons or fermions respectively.